Research
We study developmental rules and synthetically reconstruct tissue patterning
Many diseases arise from a breakdown in tissue structure, including cancers and congenital birth defects. We aim to reverse engineer the interactions between cells and their local environment that create and maintain the structure of tissues in the body.
We are particularly interested in learning to control patterning processes that occur in embryos, where the blueprint of adult tissues is established. We approach this by developing new cellular imaging, tissue construction, soft materials, and micro-scale engineering techniques.
“We aim to reverse engineer the interactions between cells and their local environment that create and maintain the structure of tissues in the body.”
Tissues that mimic morphogenesis by autonomously changing shape
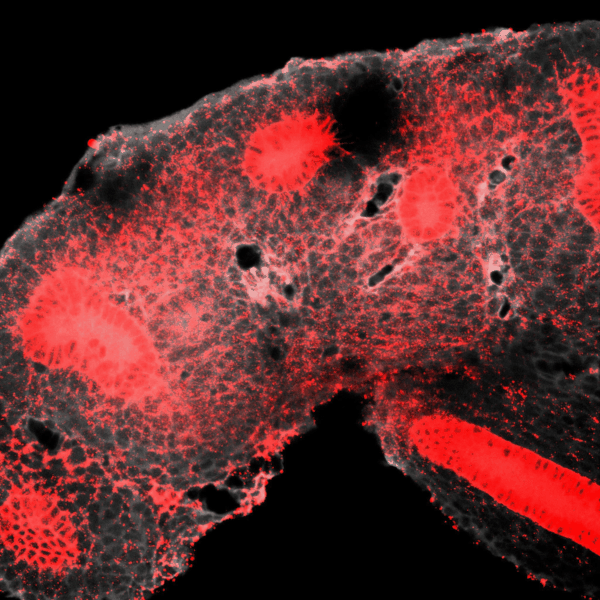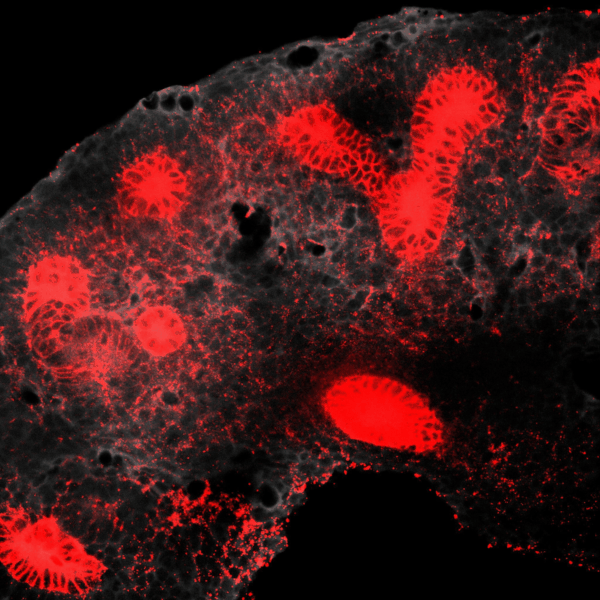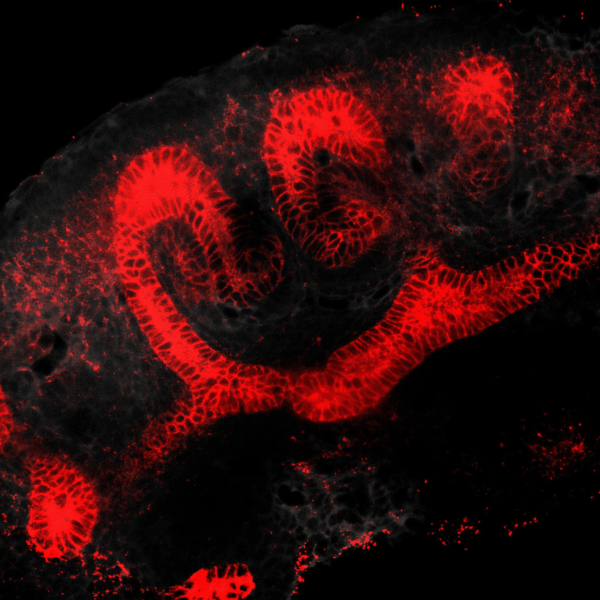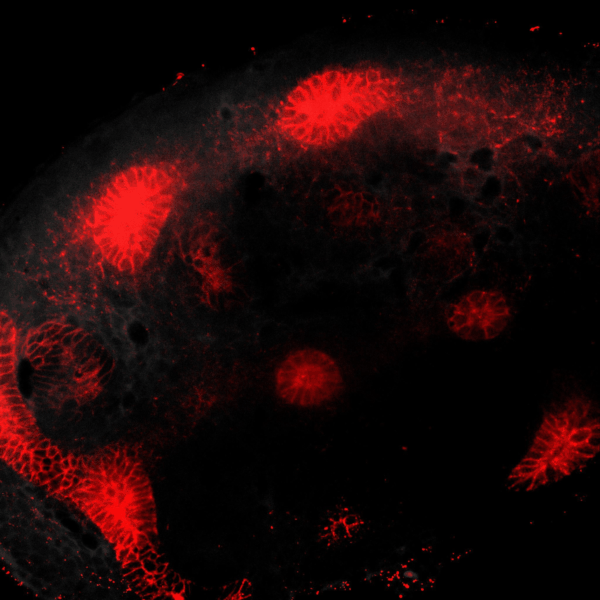
Tissues develop by progressively increasing their complexity in shape and cellular composition. We produce synthetic tissues that mimic these processes by controlling the geometry of strains generated by cells within extracellular matrix-mimetic hydrogels. We are using these fully biological shape-shifting scaffolds to study how tissue development is coordinated during, for example, branching morphogenesis of the kidney collecting duct network. We also study how mechanics and biochemical factors such as morphogens are integrated in embryonic tissue-building processes.
BRANCHING MORPHOGENESIS AND COLLECTIVE CELL MIGRATION
The kidney is a collection of blood-filtering nephrons that are interconnected through a highly organized network of epithelial tubules. The epithelial network is formed during embryonic development by the invasion and branching of epithelial progenitor tissue into early mesenchymal tissue. Defects in its organization can result in improper transport or elimination of urine. We are engineering 3D culture systems to understand the mechanical and biochemical signals kidney epithelial progenitors sense and interpret to proliferate, invade stromal tissue, and undergo branching morphogenesis. Specifically, following perturbation of kidney epithelial tissue shape or genetics, stromal cell composition, or extracellular matrix properties, we use live confocal microscopy to record and analyze epithelial collective migration outcomes.
ENGINEERING PHYSICOCHEMICAL CONTROL FOR ORGANOID MODELS
Pluripotent stem cells transition through stereotyped stages of growth, differentiation, and morphogenesis to build complex tissues and organ systems. During the development of organs that contain repetitive, functional units, such as the kidney, these processes are faithfully iterated thousands of times to form spatially organized networks. However, in vitro, initial stochastic differences in stem cells and culture conditions contribute to unintended variation within organoid models. By engineering enhanced physicochemical control over culture microenvironments, we aim to guide multicellular self-organization toward desired and predictable outcomes. These efforts will increase the throughput and robustness of organoid production, ultimately improving organoid application in drug/genetic screens and regenerative medicine.
CELL-CELL SIGNALING AT TISSUE INTERFACES
Developmental signaling pathways must be tightly controlled in space and time to pattern complex organ structures and avoid defects. For example, the embryonic kidney emerges from a simple epithelial bud to form an arborized collecting duct network through reciprocal signaling interactions with the surrounding mesenchyme. Using DNA-based cell patterning technology, we synthetically reconstruct these cell interfaces within engineered tissue scaffolds. We then use live imaging and mathematical modeling to track and study cellular interactions in space and time.
UNveiling CELL HETEROGENEITY THROUGH PROTEOMICS
Cell functional states can change rapidly, especially during development and differentiation. Assessing cell functional heterogeneity represents a significant technological challenge that requires single-cell analysis of protein levels and post-translational modifications, which execute cell functions. However, most information about individual cell states is currently gleaned through transcriptional analysis, such as single cell RNA sequencing. While low signals can be detected, transcriptional information represents an incomplete snapshot of cell state. We aim to bridge this knowledge gap with novel proteomics tools to understand cell function and disease progression.